Section: New Software and Platforms
Magus: Genome exploration and analysis
Participants : David James Sherman [correspondant] , Pascal Durrens, Florian Lajus, Xavier Calcas.
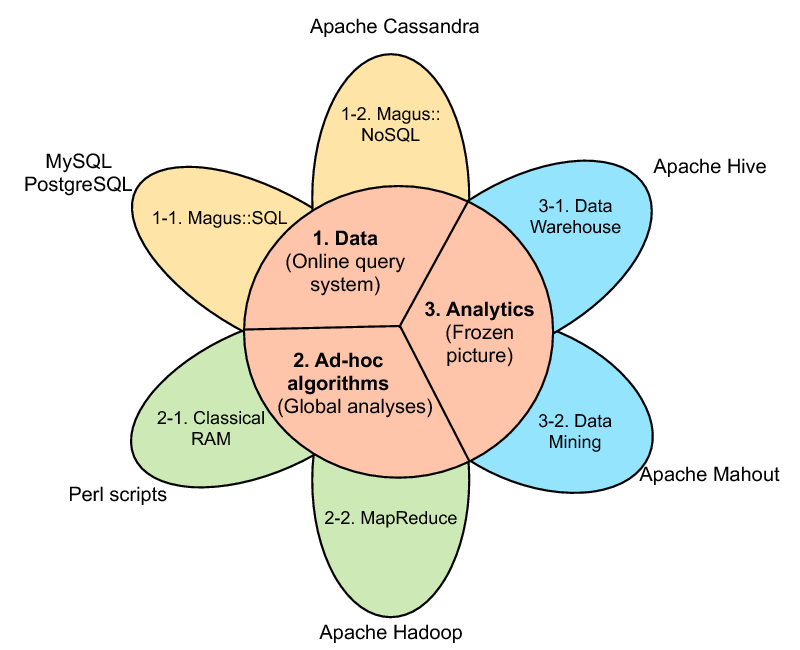
The MAGUS genome annotation system integrates genome sequences and sequences features, in silico analyses, and views of external data resources into a familiar user interface requiring only a Web navigator. MAGUS implements annotation workflows and enforces curation standards to guarantee consistency and integrity. As a novel feature the system provides a workflow for simultaneous annotation of related genomes through the use of protein families identified by in silico analyses; this results in an -fold increase in curation speed, compared to curation of individual genes. This allows us to maintain standards of high-quality manual annotation while efficiently using the time of volunteer curators. MAGUS can be used on small installations with a web server and a relational database on a single machine, or scaled out in clusters or elastic clouds using Apache Cassandra for NoSQL data storage and Apache Hadoop for Map-Reduce (figure 1 ). For more information see the MAGUS Gforge web site. (http://magus.gforge.inria.fr ) MAGUS 2.0 was developed in an Inria Technology Development Action (ADT) and is distributed with an open-source license.